Reconstructing large interaction networks from empirical time series data
Chun-Wei Chang and Chih-hao Hsieh
Prof. Chih-hao Hsieh from the Institute of Oceanography and postdoc Dr. Chun-Wei Chang from the National Center for Theoretical Science, NTU lead a research team and developed a novel method that reconstructs large interaction networks using empirical time series data. This study, published in Ecology Letters (Oct, 2021), overcomes the long-lasting challenge of reconstructing high-dimensional interaction networks in natural environments based on empirical observations. The proposed method provides a powerful tool to investigate natural time-varying interaction networks composed of a large number of interacting components.
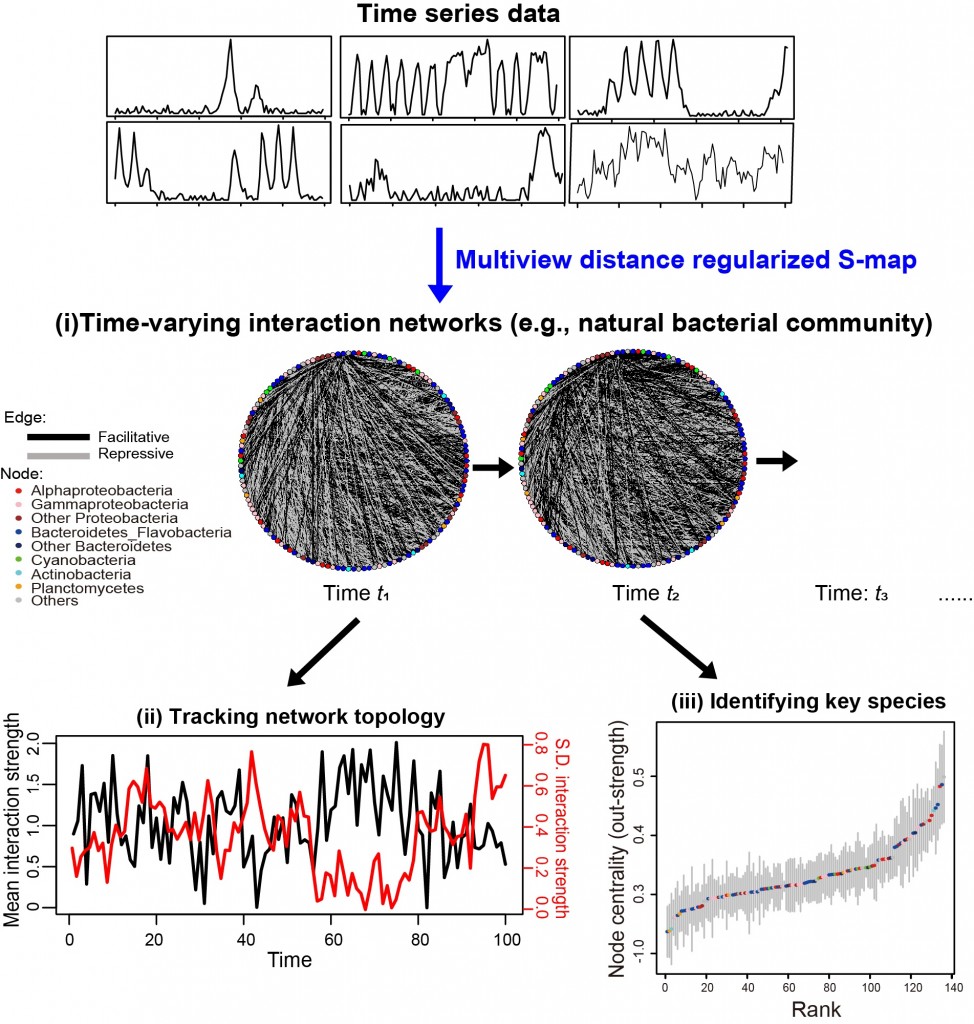
Most of ecological networks, such as food webs, mutualistic, and competition network, are high-dimensional and composed of numerous time-varying interactions among a large number of biological species, especially in those systems with high biodiversity (e.g., microbial communities, coral reefs, and rain forests). It is well-known that detailed information of network structure is needed to evaluate dynamic stability and resilience of natural systems. However, existing approaches for network construction are only applicable to small networks. Here, the research team developed a novel analytical method, multiview distance regularized S-map (MDR S-map), that overcomes “Curse of dimensionality” and reconstructs large interaction networks with time-varying interaction strengths, using time series data alone. The reconstructed networks faithfully preserved various key network properties, such as distribution of interaction strength, node centrality, and dynamical stability in numerous model systems. Applying the approach on natural bacterial communities (with 135 dominant species) tracked the temporal variations of key network topological properties and identified the most influential bacterial species occupying central positions in the network (Fig. 1).
The proposed method fulfills the public needs for reconstructing large, time-varying biological networks, especially in the field of ecology that strives to disentangle complicated interaction networks among enormous numbers of species. This method can be generalized to other types of biological networks (e.g., protein, metabolic, or neural networks) as well as non-biological networks (e.g., networks of financial trades or disease), given sufficient time series data are available.
Figure 1. The proposed network reconstruction method, MDR S-map, enables to reconstruct high-dimensional interaction networks among numerous species (i.e., network nodes) using time series data. Here is an example based on the analysis of natural bacterial communities collected in a coastal environment. The reconstructed interaction networks are time-varying and allows to track the temporal variations in network topology and to identify the key species occupying the most central positions of interaction networks.
Reference:
Chang, C. W., T. Miki, M. Ushio, P. J. Ke, H. P. Lu, F. K. Shiah, C. h. Hsieh. (2021) Reconstructing large interaction networks from empirical time series data. Ecology Letters. https://doi.org/10.1111/ele.13897